Inflammatory bowel disease (IBD), encompassing ulcerative colitis (UC), Crohn’s disease (CD) and IBD-Undefined, is a chronic inflammatory disorder of the gastrointestinal tract (1). Globally, this disease impacts over six million people (2), with approximately 25% of patients developing the disease before age 20 (1).
Living with IBD involves managing chronic symptoms and navigating periods of active disease and flare-ups, typically followed by remissions that can last months or even years (3). Recurrent episodes often require hospitalization, and many patients will need at least one operation in their lifetime, greatly affecting quality of life (4).
The concept of precision medicine—where patients are classified in subpopulations according to their clinical and molecular characteristics so that treatment strategies can be tailored to those characteristics—remains an unmet need in IBD (5). IBD is a complex disease with substantial patient heterogeneity (patients vary tremendously in disease location, behavior, and severity), which is why precision medicine is so challenging (6). Nonetheless, precision medicine could have a profound impact in diagnosis, disease course prediction, and treatment response prediction for patients with IBD (7).
This article will explore these three applications for precision medicine in-depth, detailing current approaches, recent advancements, and future opportunities. We’ll also shed light on how omics data can drive us faster towards the clinical use of IBD biomarkers for these use cases.
IBD Diagnosis: Current Approaches and Opportunities
There is no universally adopted standard for IBD clinical diagnosis. Clinicians still depend on a combination of reported symptoms (including diarrhea, abdominal cramping, nausea, pain, rectal bleeding, and fever), various blood and stool tests, radiology scans, endoscopic procedures, and histological evaluation of tissue biopsies (4).
This means that the diagnosis journey is uncomfortable, time-consuming, and costly for patients. This difficulty in achieving a quick and accurate diagnosis can lead to suboptimal disease management and less effective treatments (4).
Non-invasive methods, like serum C-reactive protein (CRP) and fecal calprotectin (FC) tests, have shown promise in identifying active disease (8). However, the existing tests have important limitations as the same biomarkers have been found to be present in other gastrointestinal diseases (5). CRP in particular has shown poor sensitivity (9), and there are practical concerns that hinder the widespread clinical use of stool testing (4).
As a result, there’s a pressing need for innovative, accurate diagnostic tests. To help develop these tests, researchers have begun to look for associations in the underlying biological mechanisms of patients with IBD.
The etiology of IBD remains unknown, but the single greatest risk factor of developing IBD is having a close family member with the disease, suggesting a genetic influence (1). 242 variants associated with IBD have been identified through large-scale genome-wide association studies (GWAS) and whole-genome sequencing. However, despite these and other advances, only a small percentage of heritability can be explained by these variants (10).
Omics-based biomarker discovery could play a pivotal role in enhancing our understanding of IBD. A recent large genotype association study found that three loci (NOD2, MHC, and MST1 3p21) were associated with subphenotypes of IBD, mainly disease location (e.g. ileal, colorectal, ileocolonic, upper GI, or other) (11). Exome sequencing integrated with biological knowledge has also been shown to distinguish between healthy individuals and individuals with CD (12).
The role of different pathways in driving IBD inflammation has also been better understood by transcriptomic analyses comparing healthy and diseased human ileum and colon samples. In both CD and UC, inflamed and non-inflamed tissues have been shown to have altered gene expression (13). Researchers have worked to better characterize the significance of this difference and identify the crucial element driving disease pathogenesis. A recent microarray data study showed MAPK3, NDRG1 and HLA-DRA may play major roles in the progression and development of IBD (14); another RNA-seq study identified CXCL1 expression associated with significantly increased CD activity (15). This type of specificity offers the potential to create better, targeted diagnostics.
IBD Disease Course Prediction: Current Approaches and Opportunities
IBD is known for its unpredictability of disease course and outcomes, which contributes significantly to disease burden and has a large impact on quality of life. When a patient is diagnosed with IBD, it’s difficult to tell them what to expect for long-term disease trajectory (i.e., whether the disease will follow a slow, lethargic course, or a more aggressive course) and when relapses or progression may occur. This makes the disease particularly difficult for patients to bear both physically and psychologically, while also making it challenging for clinicians to implement management strategies (16).
The potential application of omics biomarkers to predict disease course is promising. The field has recently started looking beyond clinical symptoms (such as gastrointestinal lesions, stricturing and penetrating behavior, etc.) alone to predict IBD course (17). For example, FC has been shown to predict the need for colectomy in UC (18) and hospitalization, surgery for CD (19). CRP is also used for prognostic prediction; however, both of these biomarkers have received conflicting reports on their usefulness in practice (20). An innovative study published earlier this year identified a panel of serum N-glycomic biomarkers and, using machine learning, developed a predictive model to anticipate disease flares with high accuracy (21).
CDPath (Takeda) and IBDX (LabCorp), two blood tests that can help predict the potential for developing serious CD disease-related complications, can be ordered by physicians but are not widely used in clinical practice due to the need for more definitive evidence (22).
There has also been significant progress using transcriptomics to predict disease course. In pediatric populations, researchers have used RNA-Seq data to develop a transcriptional risk score that shows positive results in identifying patients with CD who are likely to progress to complicated disease (23). This is a promising prediction model that needs further testing and validation with adult cohorts. Another encouraging development is the first commercially available test (PredictSURE) on RNA extracted from blood samples which shows promising initial results. The test uses gene expression data as a predictor of more aggressive disease (24).
Despite the immense progress in defining genomic, transcriptomic, and even gut microbial signatures (25) to predict disease course, none of these approaches have been definitively shown to improve clinical outcomes yet. Ultimately, more research is needed (26). With a clear tool for disease course prediction, clinicians could adopt a more proactive versus reactive strategy for treatment and intervention, resulting in an improved quality of life for patients.
IBD Treatment Response Prediction: Current Approaches and Opportunities
Common treatments for IBD episodes vary in intensity, from aminosalicylates (5-ASAs) for mild-to-moderate cases to corticosteroids and immunomodulators for moderate-to-severe situations. Inadequate response to these treatments leads to biologic therapies such as anti-tumor necrosis factor (anti-TNF) therapies and small molecular Janus kinase (JAK) inhibitor therapies. Patients may also be started on biologic therapies if their disease is severe at diagnosis (3).
Biologic therapies have significantly improved disease outcome and quality of life for IBD patients with established disease (27). These biologic therapies have been so successful that biosimilars began hitting the market this year, adding competition to major blockbusters such as AbbVie’s Humira (28).
Despite an expanding range of effective treatments for managing IBD symptoms, we urgently need biomarkers to tailor these therapies to individual patients and optimize their use (7). A significant number of patients do not respond to biologic treatments, their responsiveness diminishes over time, or they experience intolerances (29). It’s not uncommon for patients to initially show positive responses to therapeutics, but quickly develop a tolerance, forcing them to cycle through different drugs in the same class before progressing to more aggressive treatments or resorting to surgery. Non-responders not only suffer from progressive tissue damage as their disease continues to worsen without proper treatment (29), but an ineffective treatment can also influence their response to future therapies. The first biologic, regardless of choice, has the highest chance of success, and the sequence of drugs can also impact the effectiveness of a therapy (30).
Due to this dynamic, clinicians are increasingly relying on biology rather than clinical symptoms to tailor aggressive treatments for those predicted to have severe disease progression, while avoiding potent therapies for those predicted to have a benign disease course at diagnosis (17) (Figure 1). Biomarkers could be used to accurately predict IBD patients who are likely to respond well to currently available treatments. Early intervention with a therapy tailored to the patient’s biology could improve disease management and clinical outcomes (31), avoid unnecessary side effects—including the emotional toll associated with treatment failure (4)—reduce healthcare expenses (32), and even improve clinical trial recruitment (7).
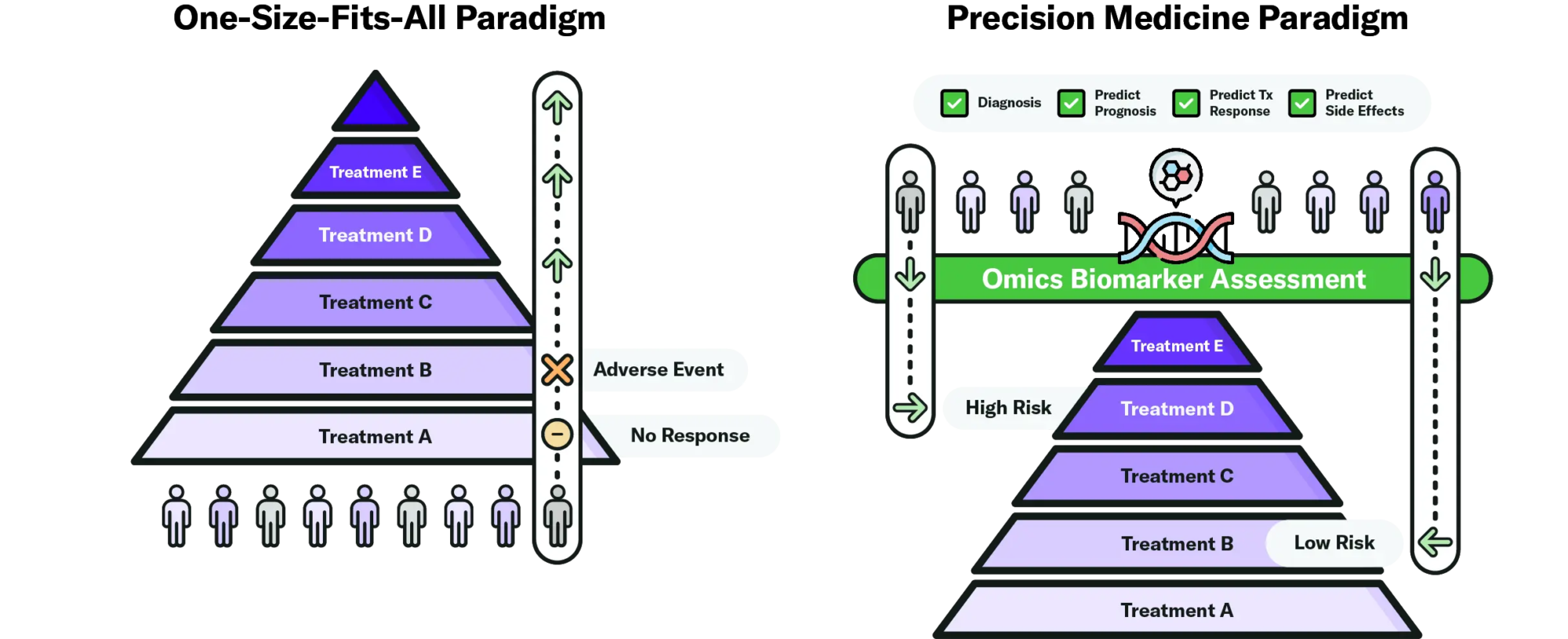
Figure 1. Omics biomarkers could be used to further optimize the treatment strategies for IBD therapies that are already commercially available. Instead of waiting for the patient’s response to treatment to better understand their disease and escalate to the next treatment, clinicians could use omics biomarkers to more accurately diagnose the patient, predict their prognosis and treatment response. That allows them to tailor treatment strategies, starting high-risk patients on aggressive therapies that will lead to better disease outcomes and reducing exposure to unnecessary and costly biologics for low-risk patients. Current clinical practice is closer to the paradigm on the left (26).
Currently, despite a significant number of potential biomarker candidates and supporting scientific data, there are no clinically available biomarkers that accurately predict treatment response to IBD therapies (33). However, there has been substantial progress recently. For instance, a study showed that microRNA signatures in colonic biopsies of acute severe UC could classify responders and non-responders to corticosteroids, infliximab and cyclosporine (34). In CD, a 5-gene signature was found to distinguish responders from non-responders to infliximab (35). In addition, whole blood and mucosal TREM1 expression have reliably indicated a poor response to anti-TNF therapy (36). This is particularly compelling as blood markers are more patient-friendly.
Evidence also suggests that the success of a drug may depend on the state of the disease when the drug is administered. Perhaps some patients do not respond because their temporary disease inflammation is too severe for the drug to mitigate. Further studies to validate this hypothesis—for example, by performing transcriptomics on mucosal tissue samples—could lead us to a novel predictive marker for inflammation that is specific to disease state (7). Precision medicine could also predict treatment side effects, which could be used to guide treatment choice. A recent study, for instance, identified a marker within the HLA region associated with 5-ASA-induced nephrotoxicity (37). This information could be used to tailor renal function monitoring strategies in patients taking 5-ASA agents.
The progress described above demonstrates the immense potential for omics signatures to provide clinicians with prognostic information to make better informed treatment decisions. However, further validation of these biomarkers is required before they can be implemented in clinical practice (26).
How Omics Data Will Help Accelerate Clinical Use of IBD Biomarkers
Though many potentially life-changing IBD biomarkers have been identified, significant challenges remain in translating this research to clinical practice (38). We still lack standardization and harmonization regarding the selection and cost-effectiveness of these biomarkers, and whether they indeed improve patient outcomes (7).
The heterogeneity of IBD and lack of validation studies are two of the main reasons cited as barriers to applying biomarkers to clinical practice. Nonetheless, there is much optimism that the integration of omics data into IBD research will allow for deep analysis that increases our understanding of complex traits and unlocks a new dimension in biomarker discovery (38). Here, we discuss three main ways in which omics data can continue to propel IBD discoveries forward.
Applying machine learning models to integrated multi-omics datasets
The scientific community generally believes that IBD is the result of a combination of not only genetic and environmental factors, but also microbial and immunologic factors (17). Therefore, it makes sense for researchers to take a systems biology approach by collecting diverse data types (genomics, transcriptomics, etc.) from multiple sources and integrating them, rather than continuing to search on single factors (38).
A recent standout omics data study created an algorithm based on previously identified causal gene relationships and applied it to differential gene expression and clinical data to predict response to infliximab in UC patients. The authors identified TNF, interferon gamma (IFNG), and lipopolysaccharide (LPS) as potential regulators of infliximab response with an accuracy of seventy percent (39). By concurrently screening a patient’s molecular data such as genome and transcriptome, alongside their clinical data such as medical records, machine learning algorithms can identify patterns previously impossible to discern (38).
Generating richer omics datasets with advanced sequencing
With advancements in sequencing technologies, more comprehensive, in-depth omics screenings are possible, increasing the prospects for novel discoveries. Whole genome sequencing (WGS) offers advantages over genotyping arrays in understanding the role of rare variants, and has uncovered rare genetic effects for other complex diseases such as type 2 diabetes.
Recent scientific work applied WGS and whole exome sequencing (WES) to discover that a NOX1 variant contributes to very early-onset IBD (40). Another study performed WGS on samples from African-American populations, revealing an association between CALB2 variant and ulcerative colitis (41). The authors call for additional research to determine the extent to which this variant explains differences in disease susceptibility. The steadily decreasing cost of sequencing makes such studies increasingly feasible.
Investigating multi-omics datasets derived from underrepresented populations
Generally, IBD genetic studies have disproportionately focused on individuals of European ancestry, despite the disease’s prevalence across diverse populations (42). The utility of findings derived from a homogenous population is inherently limited, but this is especially the case for models of disease susceptibility, which have been demonstrated to perform significantly worse when applied to different populations (43). A recent study supporting this theory found that IBD risk prediction in African-American cohorts was less accurate than in other ancestry populations, due to small sample sizes of African ancestry in IBD association studies (42).
Retrospective omics datasets drawn from diverse patient populations can help propel pre-clinical discoveries, which can help derisk early-phase clinical trials. This is especially important for IBD as patient recruitment for placebo-controlled clinical trials is challenged by the existence of effective treatment options and restrictive eligibility criteria (44). Pre-clinical findings can bring critical decision points forward so that enrolled participants can be utilized effectively.
The Road Ahead
Taking a systems biology approach and applying machine learning models to integrated multi-omics datasets will enable researchers to capture complex associations and increase our understanding of IBD. This can build upon the commendable work already undertaken to identify biomarkers for disease diagnosis, prediction of disease course, and treatment response, thus facilitating their translation into routine clinical practice in the years to come.
Ovation’s IBD Omics Data includes WGS and RNA-Seq data with up to 30x coverage linked with relevant clinical data including pathology reports, disease severity, comorbidity diagnoses, therapeutic exposures, medical procedures, and surgical history. Researchers are already accessing this data quickly, utilizing sophisticated analytical models to discover clinically translatable knowledge. Additionally, we have the capability to rapidly generate new omics datasets by sequencing samples from our extensive biobank which comprises over 1.53 million consented samples.
Too many people live with the chronic condition of IBD, a reality made even more challenging due to the often lengthy and complex journey of diagnostics and treatments that fall short of their full potential. Ovation aspires to increase access to comprehensive omics data to accelerate the integration of precision medicine into routine clinical practice—so that patients with IBD receive the targeted diagnosis and treatment they rightfully deserve.
References
- Borowitz SM. The epidemiology of inflammatory bowel disease: Clues to pathogenesis? Front Pediatr. 2023;10:1103713. doi:10.3389/fped.2022.1103713
- Alatab S, Sepanlou SG, Ikuta K, et al. The global, regional, and national burden of inflammatory bowel disease in 195 countries and territories, 1990–2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet Gastroenterol Hepatol. 2020;5(1):17-30. doi:10.1016/S2468-1253(19)30333-4
- Crohn’s & Colitis Foundation. Understanding IBD Medications [brochure]. 2022. https://www.crohnscolitisfoundation.org/sites/default/files/2022-12/understanding-ibd-medications-brochure-final-rev0822-3_2.pdf. Accessed July 7, 2023
- Chen P, Zhou G, Lin J, et al. Serum Biomarkers for Inflammatory Bowel Disease. Front Med (Lausanne). 2020;7:123. Published 2020 Apr 22. doi:10.3389/fmed.2020.00123
- Noor NM, Verstockt B, Parkes M, Lee JC. Personalised medicine in Crohn’s disease. Lancet Gastroenterol Hepatol. 2020;5(1):80-92. doi:10.1016/S2468-1253(19)30340-1
- Fiocchi C, Dragoni G, Iliopoulos D, et al. Results of the Seventh Scientific Workshop of ECCO: Precision Medicine in IBD-What, Why, and How. J Crohns Colitis. 2021;15(9):1410-1430. doi:10.1093/ecco-jcc/jjab051
- Denson LA, Curran M, McGovern DPB, et al. Challenges in IBD Research: Precision Medicine. Inflamm Bowel Dis. 2019;25(Supplement_2):S31-S39. doi:10.1093/ibd/izz078
- Sands BE. Biomarkers of Inflammation in Inflammatory Bowel Disease. Gastroenterology. 2015;149(5):1275-1285.e2. doi:10.1053/j.gastro.2015.07.003
- Mosli MH, Zou G, Garg SK, et al. C-Reactive Protein, Fecal Calprotectin, and Stool Lactoferrin for Detection of Endoscopic Activity in Symptomatic Inflammatory Bowel Disease Patients: A Systematic Review and Meta-Analysis. Am J Gastroenterol. 2015;110(6):802-820. doi:10.1038/ajg.2015.120
- Umićević Mirkov M, Verstockt B, Cleynen I. Genetics of inflammatory bowel disease: beyond NOD2. Lancet Gastroenterol Hepatol. 2017;2(3):224-234. doi:10.1016/S2468-1253(16)30111-X
- Cleynen I, Boucher G, Jostins L, et al. Inherited determinants of Crohn’s disease and ulcerative colitis phenotypes: a genetic association study. Lancet. 2016;387(10014):156-167. doi:10.1016/S0140-6736(15)00465-1
- Jeong CS, Kim D. Inferring Crohn’s disease association from exome sequences by integrating biological knowledge. BMC Med Genomics. 2016;9 Suppl 1(Suppl 1):35. Published 2016 Aug 12. doi:10.1186/s12920-016-0189-2
- Montero-Meléndez T, Llor X, García-Planella E, Perretti M, Suárez A. Identification of novel predictor classifiers for inflammatory bowel disease by gene expression profiling. PLoS One. 2013;8(10):e76235. Published 2013 Oct 14. doi:10.1371/journal.pone.0076235
- Li XL, Zhou CY, Sun Y, Su ZY, Wang X, Jia EN, Zhang Q, Jiang XF, Qi WQ, Xu Y. Bioinformatic analysis of potential candidates for therapy of inflammatory bowel disease. Eur Rev Med Pharmacol Sci. 2015;19:4275-4284.
- Hong SN, Joung JG, Bae JS, et al. RNA-seq Reveals Transcriptomic Differences in Inflamed and Noninflamed Intestinal Mucosa of Crohn’s Disease Patients Compared with Normal Mucosa of Healthy Controls. Inflamm Bowel Dis. 2017;23(7):1098-1108. doi:10.1097/MIB.0000000000001066
- Hart AL, Rubin DT. Entering the Era of Disease Modification in Inflammatory Bowel Disease. Gastroenterology. 2022;162(5):1367-1369. doi:10.1053/j.gastro.2022.02.013
- Borg-Bartolo SP, Boyapati RK, Satsangi J, Kalla R. Precision medicine in inflammatory bowel disease: concept, progress and challenges. F1000Res. 2020;9:F1000 Faculty Rev-54. Published 2020 Jan 28. doi:10.12688/f1000research.20928.1
- Ho GT, Lee HM, Brydon G, et al. Fecal calprotectin predicts the clinical course of acute severe ulcerative colitis. Am J Gastroenterol. 2009;104(3):673-678. doi:10.1038/ajg.2008.119
- Kennedy NA, Jones GR, Plevris N, Patenden R, Arnott ID, Lees CW. Association Between Level of Fecal Calprotectin and Progression of Crohn’s Disease. Clin Gastroenterol Hepatol. 2019;17(11):2269-2276.e4. doi:10.1016/j.cgh.2019.02.017
- Kalla R, Kennedy NA, Ventham NT, et al. Serum Calprotectin: A Novel Diagnostic and Prognostic Marker in Inflammatory Bowel Diseases. Am J Gastroenterol. 2016;111(12):1796-1805. doi:10.1038/ajg.2016.342
- Shubhakar A, Jansen BC, Adams AT, et al. Serum N-Glycomic Biomarkers Predict Treatment Escalation in Inflammatory Bowel Disease. J Crohns Colitis. 2023;17(6):919-932. doi:10.1093/ecco-jcc/jjad012
- Crohn’s & Colitis Foundation. Diagnosing and Monitoring IBD [brochure]. 2018. Available at: https://issuu.com/ccfa1/docs/diagnosing-monitoring-ibd-brochure-final-rev062518. Accessed July 7, 2023
- Marigorta UM, Denson LA, Hyams JS, et al. Transcriptional risk scores link GWAS to eQTLs and predict complications in Crohn’s disease. Nat Genet. 2017;49(10):1517-1521. doi:10.1038/ng.3936
- Biasci D, Lee JC, Noor NM, et al. A blood-based prognostic biomarker in IBD. Gut. 2019;68:1386-1395. doi: 10.1136/gutjnl-2019-318343
- Haberman Y, Tickle TL, Dexheimer PJ, et al. Pediatric Crohn disease patients exhibit specific ileal transcriptome and microbiome signature [published correction appears in J Clin Invest. 2015 Mar 2;125(3):1363]. J Clin Invest. 2014;124(8):3617-3633. doi:10.1172/JCI75436
- Vieujean S, Louis E. Precision medicine and drug optimization in adult inflammatory bowel disease patients. Therap Adv Gastroenterol. 2023;16:17562848231173331. Published 2023 May 10. doi:10.1177/17562848231173331
- Colombel JF, Narula N, Peyrin-Biroulet L. Management Strategies to Improve Outcomes of Patients With Inflammatory Bowel Diseases. Gastroenterology. 2017;152(2):351-361.e5. doi:10.1053/j.gastro.2016.09.046
- Weintraub A. Coherus breached Humira patent settlement by striking deal with Mark Cuban’s drug firm: AbbVie. FiercePharma. Published July 7, 2023. Accessed July 7, 2023. Available at: https://www.fiercepharma.com/pharma/coherus-breached-humira-patent-settlement-striking-deal-mark-cubans-drug-firm-abbvie
- Colombel JF, Narula N, Peyrin-Biroulet L. Management Strategies to Improve Outcomes of Patients With Inflammatory Bowel Diseases. Gastroenterology. 2017;152(2):351-361.e5. doi:10.1053/j.gastro.2016.09.046
- Jossen J, Kiernan BD, Pittman N, Dubinsky MC. Anti-tumor Necrosis Factor-alpha Exposure Impacts Vedolizumab Mucosal Healing Rates in Pediatric Inflammatory Bowel Disease. J Pediatr Gastroenterol Nutr. 2020;70(3):304-309. doi:10.1097/MPG.0000000000002556
- Wang C, Baer HM, Gaya DR, Nibbs RJB, Milling S. Can molecular stratification improve the treatment of inflammatory bowel disease?. Pharmacol Res. 2019;148:104442. doi:10.1016/j.phrs.2019.104442
- van der Valk ME, Mangen MJ, Leenders M, et al. Healthcare costs of inflammatory bowel disease have shifted from hospitalisation and surgery towards anti-TNFα therapy: results from the COIN study. Gut. 2014;63(1):72-79. doi:10.1136/gutjnl-2012-303376
- Stevens TW, Matheeuwsen M, Lönnkvist MH, et al. Systematic review: predictive biomarkers of therapeutic response in inflammatory bowel disease-personalised medicine in its infancy. Aliment Pharmacol Ther. 2018;48(11-12):1213-1231. doi:10.1111/apt.15033
- Morilla I, Uzzan M, Laharie D, et al. Colonic MicroRNA Profiles, Identified by a Deep Learning Algorithm, That Predict Responses to Therapy of Patients With Acute Severe Ulcerative Colitis. Clin Gastroenterol Hepatol. 2019;17(5):905-913. doi:10.1016/j.cgh.2018.08.068
- Arijs I, Quintens R, Van Lommel L, et al. Predictive value of epithelial gene expression profiles for response to infliximab in Crohn’s disease. Inflamm Bowel Dis. 2010;16(12):2090-2098. doi:10.1002/ibd.21301
- Verstockt B, Verstockt S, Dehairs J, et al. Low TREM1 expression in whole blood predicts anti-TNF response in inflammatory bowel disease. EBioMedicine. 2019;40:733-742. doi:10.1016/j.ebiom.2019.01.027
- Heap GA, So K, Weedon M, et al. Clinical Features and HLA Association of 5-Aminosalicylate (5-ASA)-induced Nephrotoxicity in Inflammatory Bowel Disease [published correction appears in J Crohns Colitis. 2017 Dec 4;11(12):1509]. J Crohns Colitis. 2016;10(2):149-158. doi:10.1093/ecco-jcc/jjv219
- Seyed Tabib NS, Madgwick M, Sudhakar P, Verstockt B, Korcsmaros T, Vermeire S. Big data in IBD: big progress for clinical practice. Gut. 2020;69(8):1520-1532. doi:10.1136/gutjnl-2019-320065
- Zarringhalam K, Enayetallah A, Reddy P, Ziemek D. Robust clinical outcome prediction based on Bayesian analysis of transcriptional profiles and prior causal networks. Bioinformatics. 2014;30(12):i69-i77. doi:10.1093/bioinformatics/btu272
- Schwerd T, Bryant RV, Pandey S, et al. NOX1 loss-of-function genetic variants in patients with inflammatory bowel disease. Mucosal Immunol. 2018;11(2):562-574. doi:10.1038/mi.2017.74
- Somineni HK, Nagpal S, Venkateswaran S, et al. Whole-genome sequencing of African Americans implicates differential genetic architecture in inflammatory bowel disease. Am J Hum Genet. 2021;108(3):431-445. doi:10.1016/j.ajhg.2021.02.001
- Gettler K, Levantovsky R, Moscati A, et al. Common and Rare Variant Prediction and Penetrance of IBD in a Large, Multi-ethnic, Health System-based Biobank Cohort. Gastroenterology. 2021;160(5):1546-1557. doi:10.1053/j.gastro.2020.12.034
- Duncan L, Shen H, Gelaye B, et al. Analysis of polygenic risk score usage and performance in diverse human populations. Nat Commun. 2019;10(1):3328. Published 2019 Jul 25. doi:10.1038/s41467-019-11112-0
- Uzzan M, Bouhnik Y, Abreu M, et al. Declining Enrolment and Other Challenges in IBD Clinical Trials: Causes and Potential Solutions. J Crohns Colitis. 2023;17(7):1066-1078. doi:10.1093/ecco-jcc/jjad020